
ABCD STUDY: STUDY
DESIGN, DATA SHARING &
DEAP
Wesley K. Thompson | August 20, 2019

STUDY DESIGN

3
ABCD STUDY DESIGN
• The complete collection of baseline
data was released on the NIMH
Data Archive (NDA) in March 2019.
• Baseline data are assessed on
11,878 subjects at 21 sites around
the country.
• There are also follow-up
assessments on a minority of these
subjects.

ABCD data dictionary (release 2.0)
27,400 x 65,000
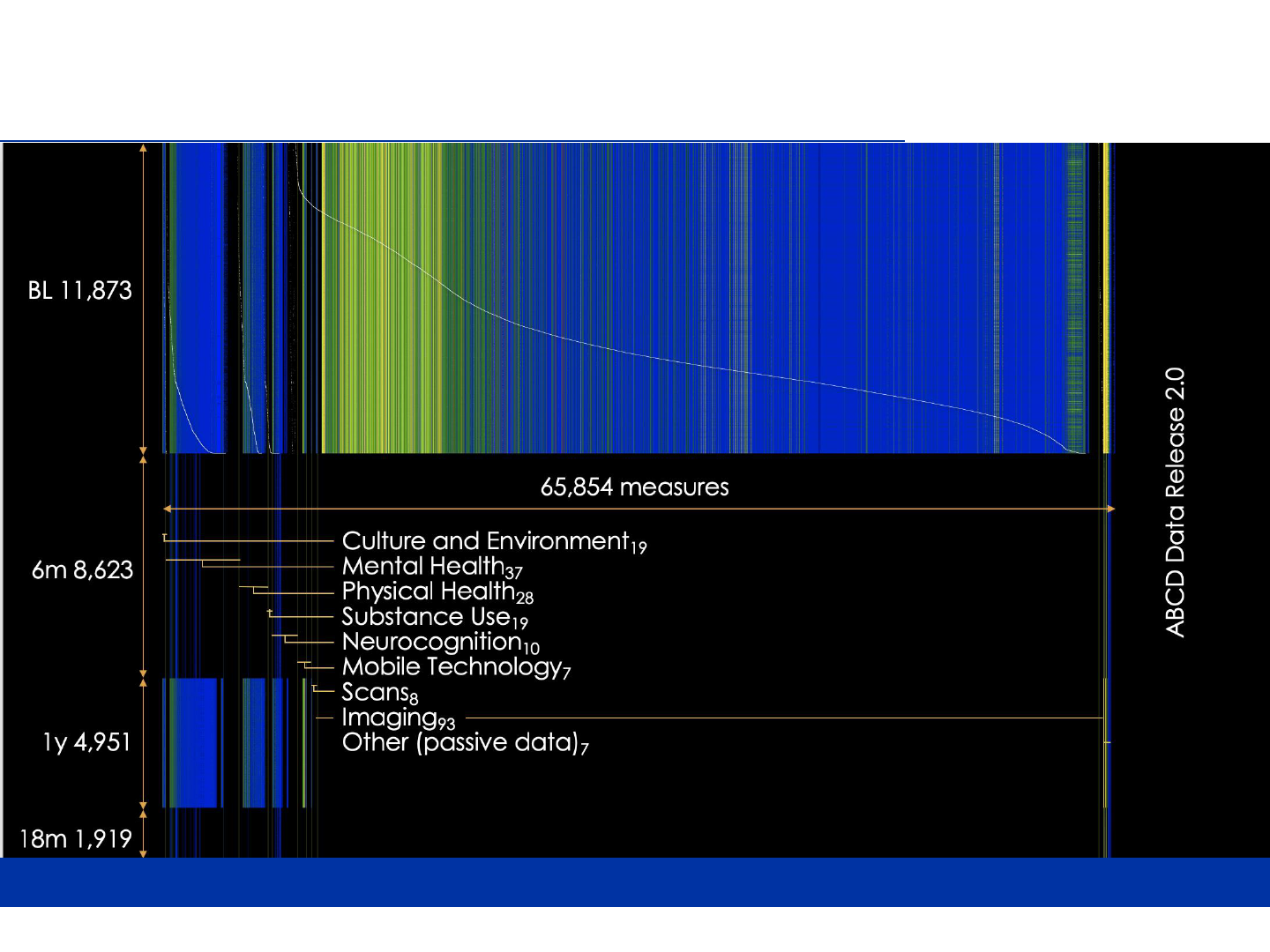
5
ABCD STUDY DESIGN (SHARED DATA IN 2.0)
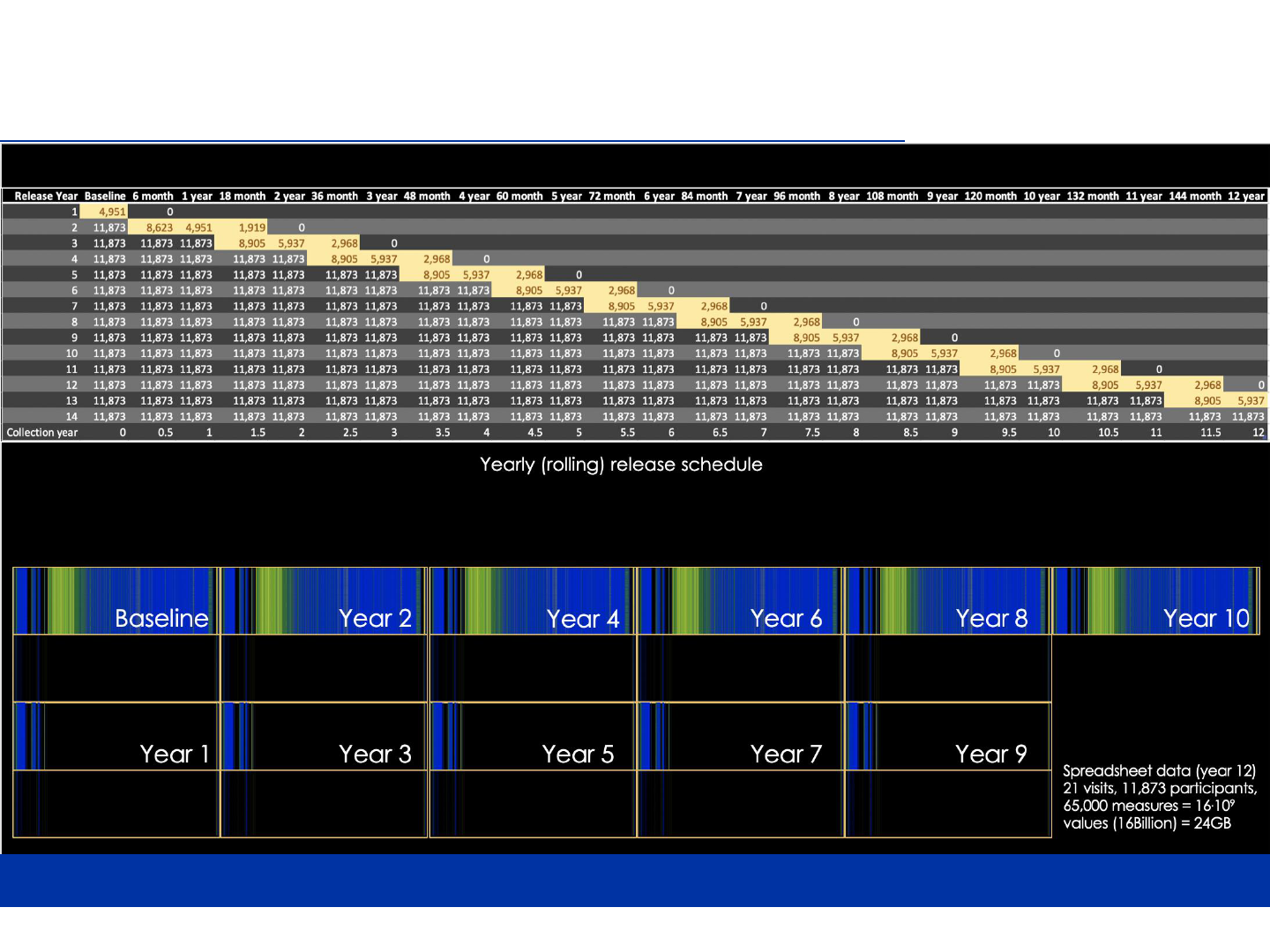
6
ABCD STUDY DESIGN – DATA RELEASE SCHEDULE

7
ABCD STUDY DESIGN

Missing data
✓ I don’t know
✓ I don’t want to tell you
✓ Truly missing
✓ Messed up, never asked
✓ Lost in transmission
✓ We have answers but no participant ID
✓ Missingness by design (not missing)
✓ By event type (e.g. no imaging data at non-imaging events)
✓ New questionnaires/Variables are introduced – missing before date
✓ Missing because of branching logic

DATA SHARING

Shared data, opportunities/challenges
• ABCD Policy: All data is shared on an ongoing basis – no holdout data.
Any results published require a pre-release of that data.
• Single channel for data release on National Data Archive.
• Share standard results such as results from QC pipelines and derived
scores is good
• lower barrier for analysis entry
• use the community to provide feedback
• promote best practices
• reduce researchers degrees of freedom
• Requires additional resources for data curation, additional
documentation, data sharing and communication towards the
community. Exposes study to more challenging events.
Harmonization of no interest
Name changes require extensive coupling
lists for quality assurance
Harmonization of value
Coding of complex data during acquisition to
allow for linkage to external information sources
A study centric view of data harmonization
Supported now by NDA:
• Alias fields in data dictionary
• Study specific download packages
Supported by ABCD:
• Use of RxNorm for medication inventory
• Use of consistent names for brain ROIs
DEAP applications for specialized domains
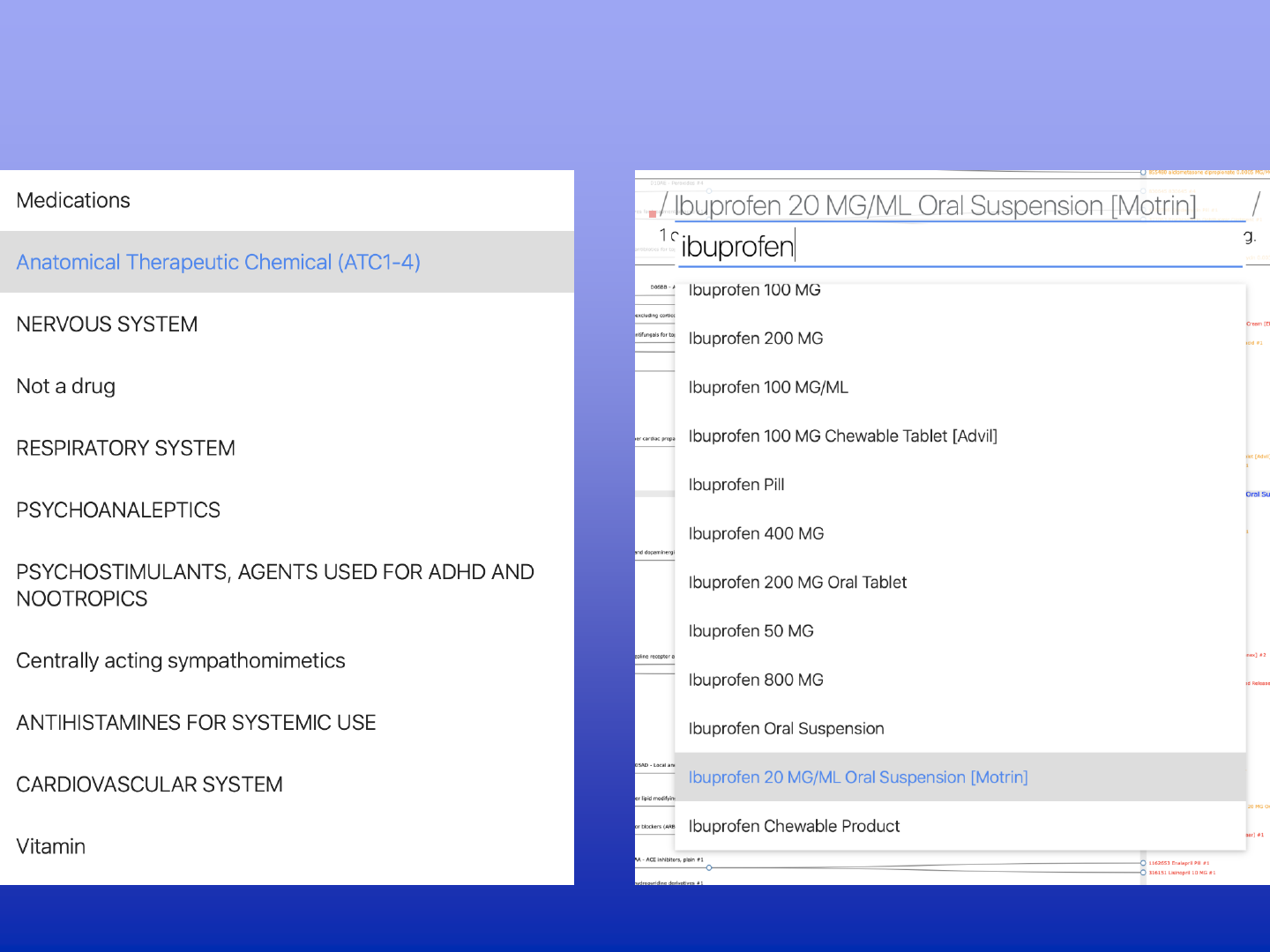

DATA EXPLORATION AND
ANALYSIS PORTAL (DEAP)

Data Exploration and Analysis Portal
Web-based interface, cloud deployment
NIMH’s NDA data sharing platform as data source
Access to all ABCD measures shared in NDA17
Build-in nesting for multi-level covariates of choice
Access to visualizations and statistical model summary

Shared ABCD data
Available on National Data Archive (nda.nih.gov)
requires signup and support from institution
11,875 participants data available since early 2019
3.2GB spreadsheet data (*.tsv)
23TB MRI (300Gb T1/T2)
65,000 measures per participant
(>67% from imaging)
Resources:
Source code repositories - github.com/ABCD-STUDY/
Data Analysis and Exploration Portal
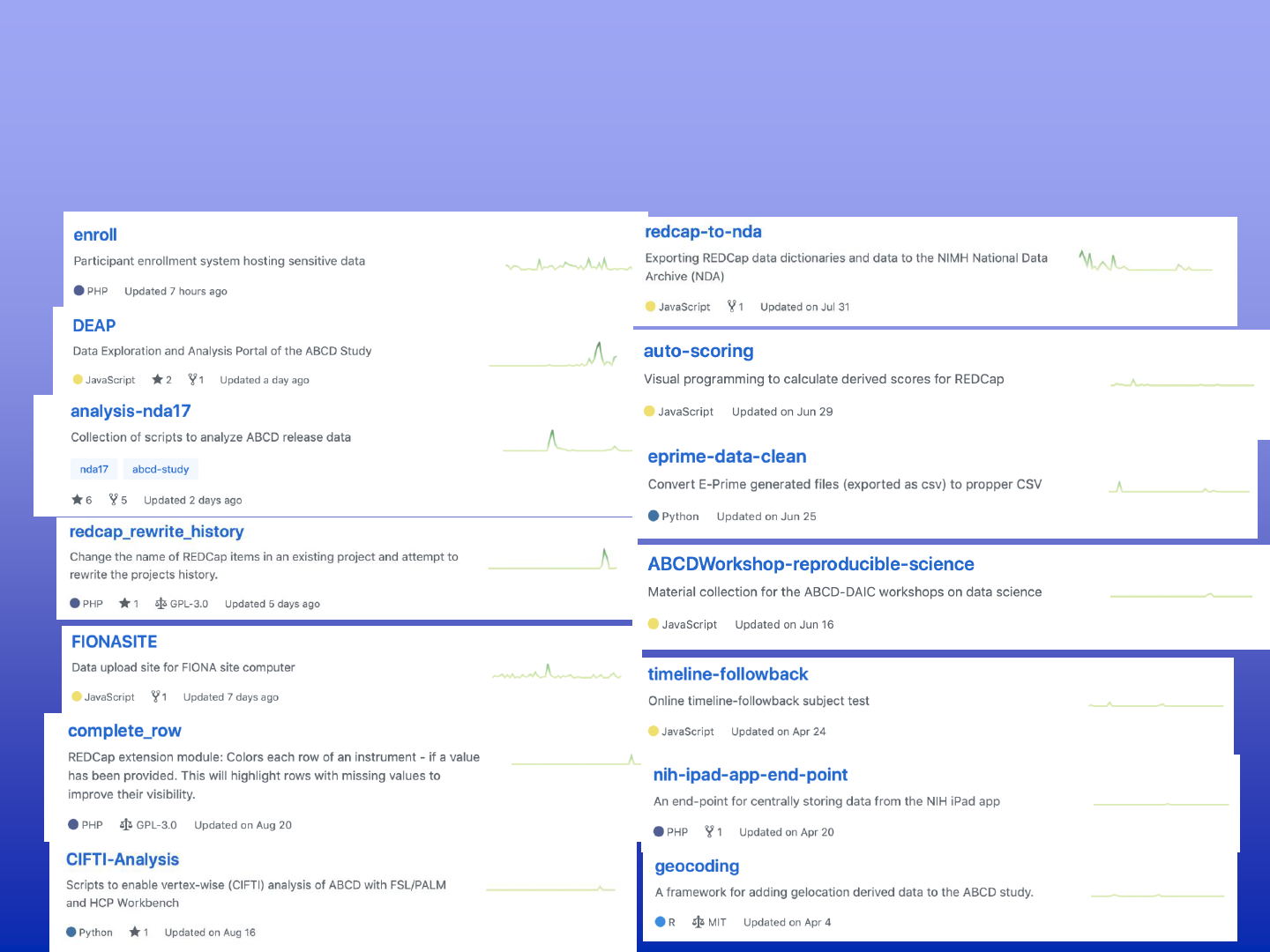
ABCD open science
[1 Team, 15 members, 33 git repositories]

DEAP web-interface
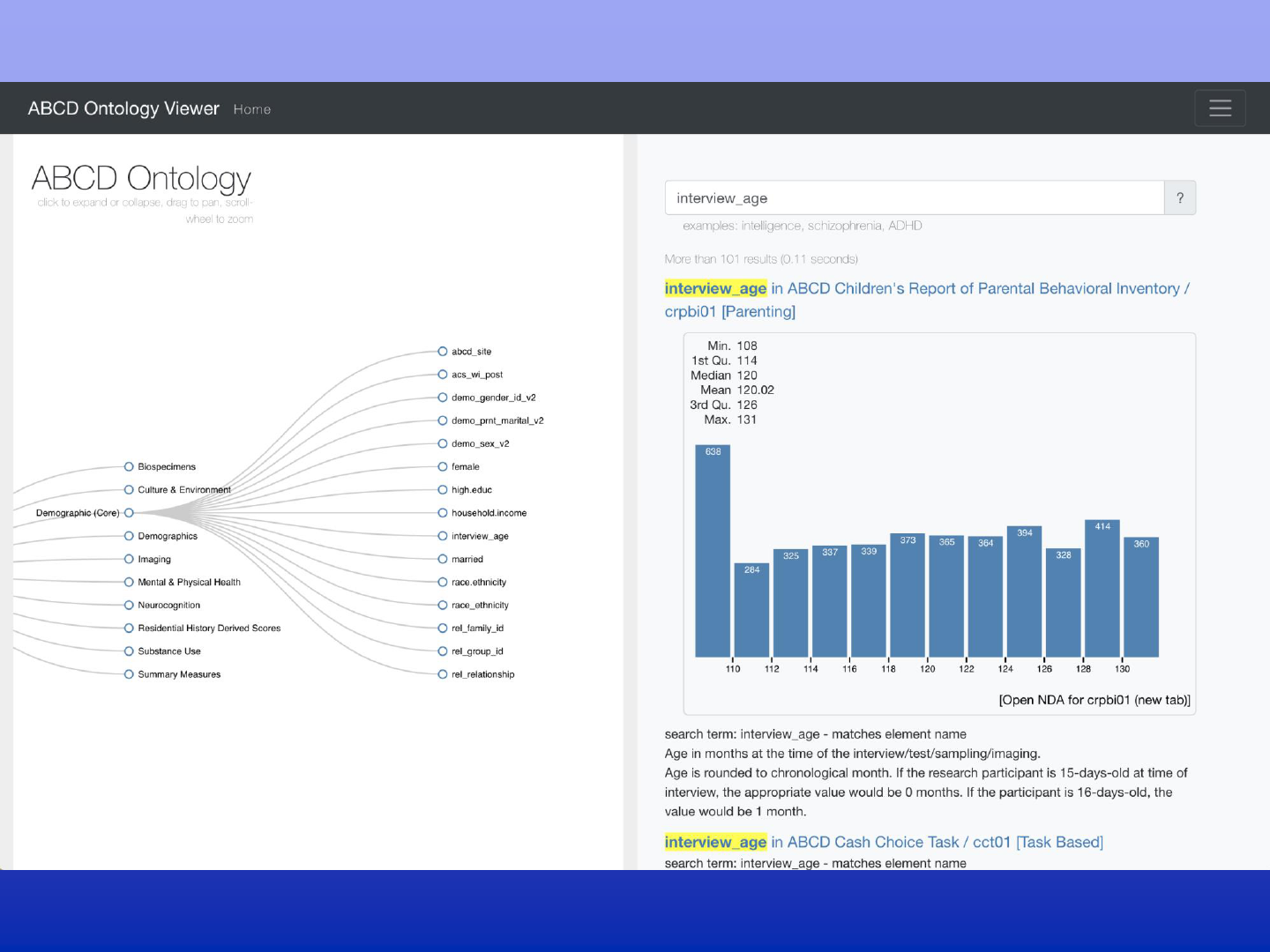
Explore 44,000 ABCD measures

Visual sub-setting data exploration
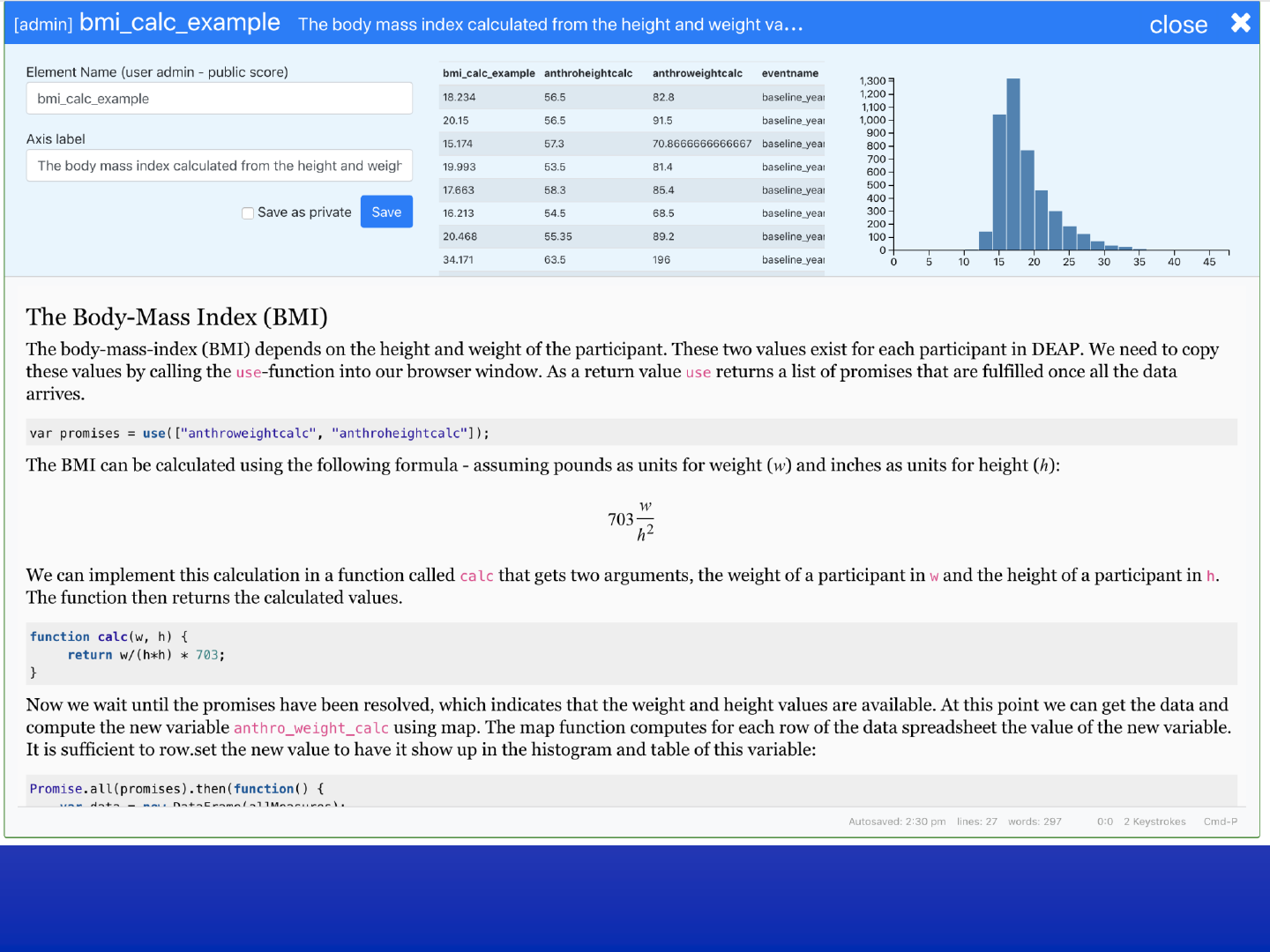
Notebook style, user defined derived measures

Multilevel Data Analysis
Multilevel statistical models for baseline data reflect the
multilevel study design (GAMM4).
• x
sfi
are covariates (e.g., demographics)
• z
sfi
are independent variables of interest
• a
s
is a site-specific random effect
• b
f(s)
is a family random effect nested within site
This model is extendable to non-normal outcomes
(e.g., discrete, count variables).

24
ABCD STUDY DESIGN
• Of these 11,875 subjects, family units include:
• 8,150 singletons
• 1,600 non-twin siblings
• 2,100 twins (1,050 pairs)
• 30 triplets (10 sets)
25
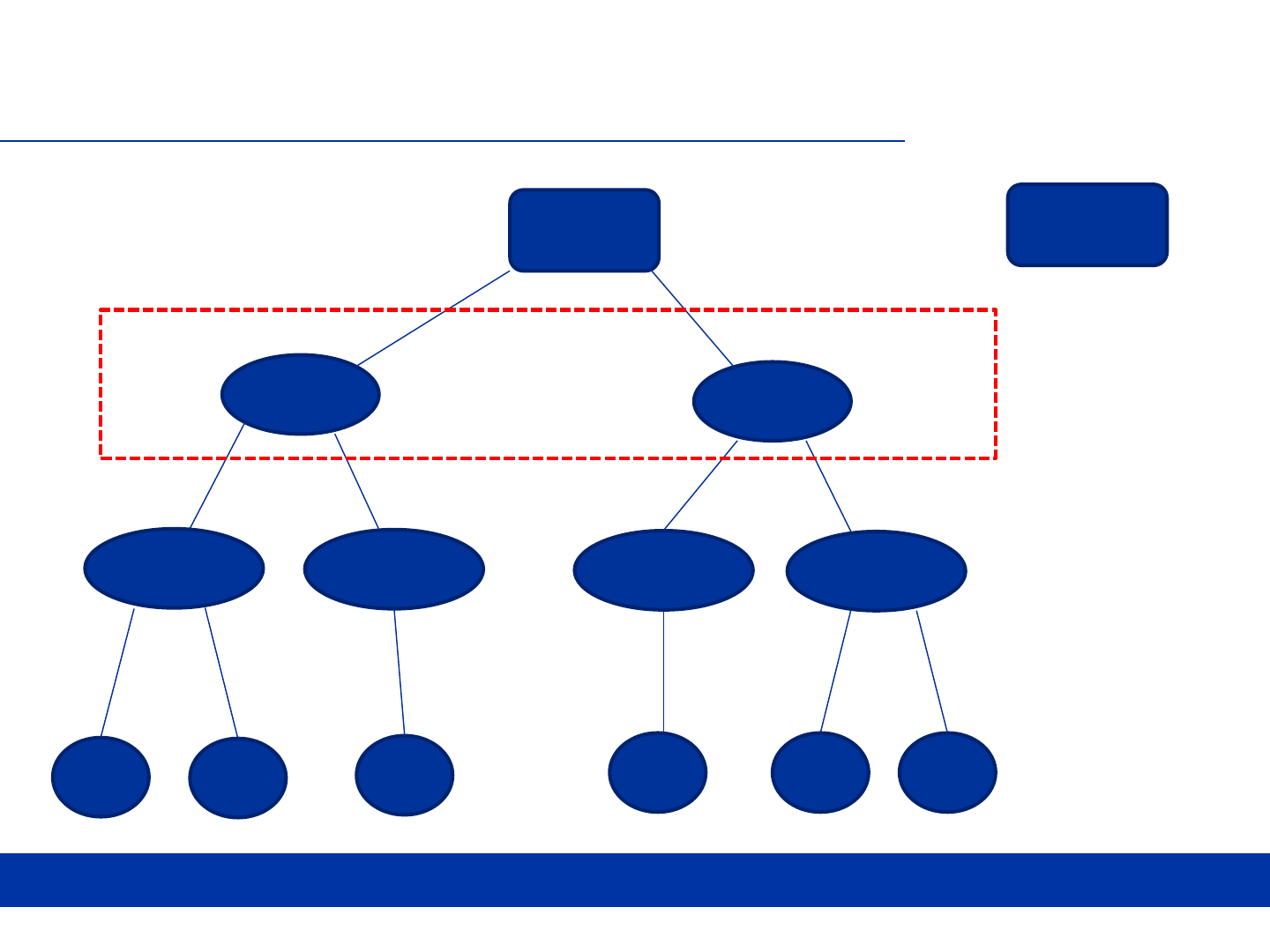
ABCD STUDY DESIGN
• …
Site 1
Site 21
MR 1
MR 2
Fam 1
Fam 2
Fam 3
Fam 4
S1
S2
S3
S4 S5 S6
Tutorial Mode on DEAP
Not familiar with generalized additive mixed models for the analysis of longitudinal data in a
multi-site project with a complex family structure? Deap provides a training-wheel mode with
in-depth explanations on how to interpret your model.
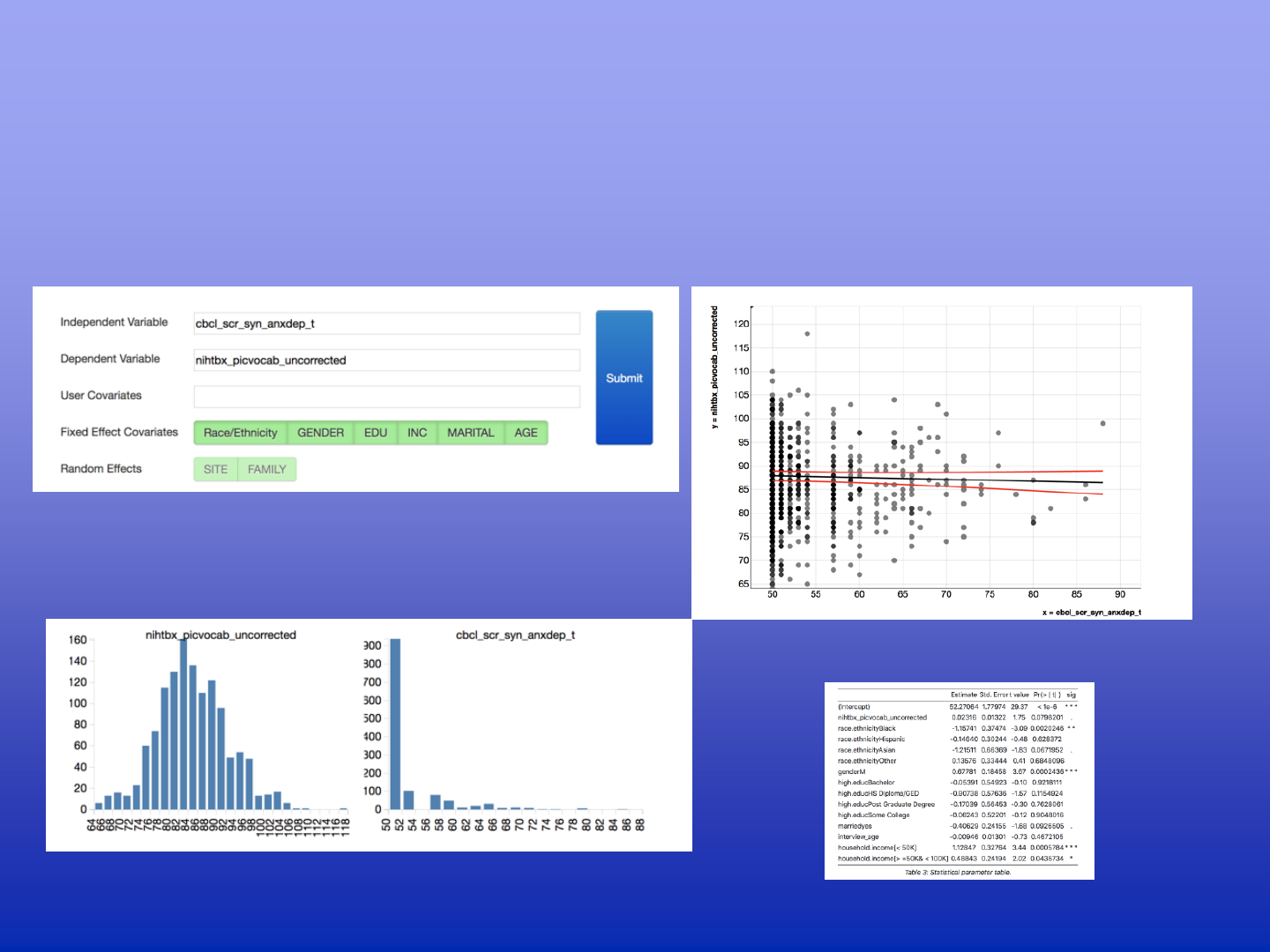
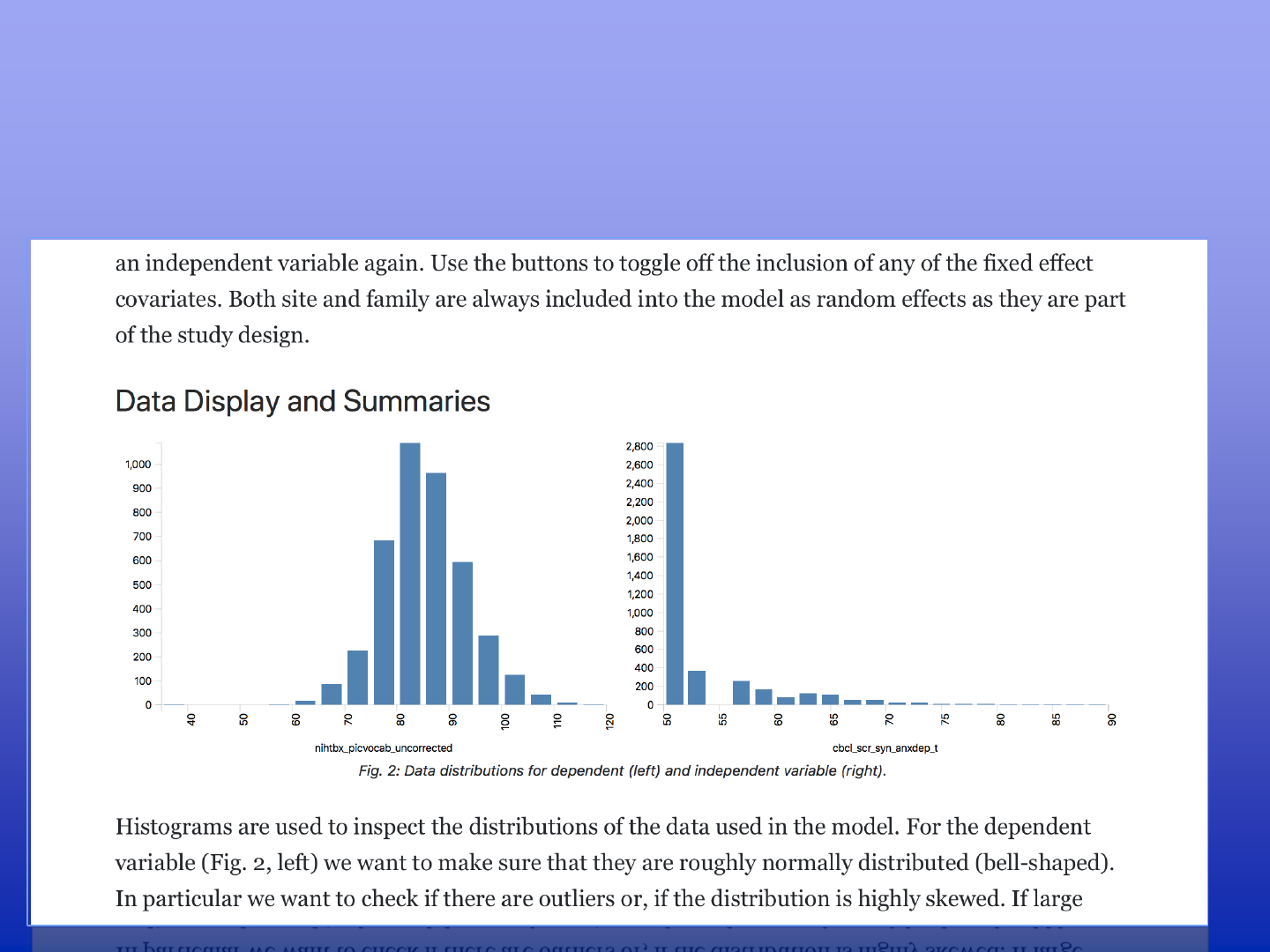
Hypothesis Testing on DEAP
Can changes in anxiety be explained by cognitive development scores measured
in the picture vocabulary test, if one corrects for known covariates?
A Model specification
B Data used in the model
C Regression model fit
D Result tables / Model comparisons
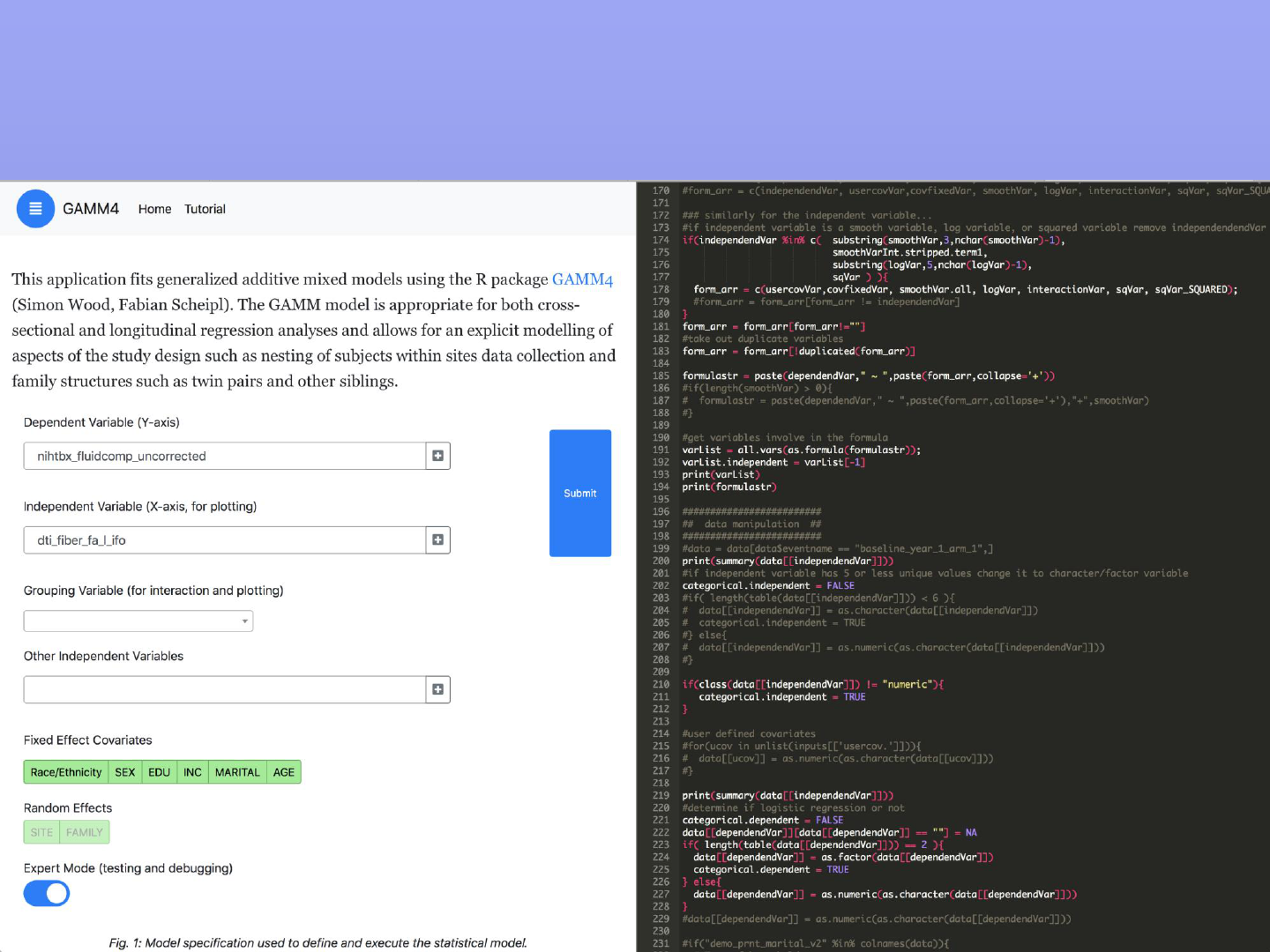
Feature: Expert Mode
Access to the (R) source code behind the GAMM4 model. Can be edited by the user and
becomes part of a sharable resource for download and to other DEAP users.

DEAP Updates
• Docker deployment of DEAP (github.com/ABCD-STUDY/DEAP).
• Pre-registration workflow supporting model specification with variable
selection and appropriate variable transformations. Text is provided for
sampling, design, and analysis plan as well as for the analysis scripts.
• Subset analysis of participants.
• User defined derived variables with data dictionary entries and scoring
algorithms (sharable).
• Upcoming:
• Allow for
– additional projects shared on DEAP (NDA17, NDA18),
– additional participants (add to our replace ABCD cohort)
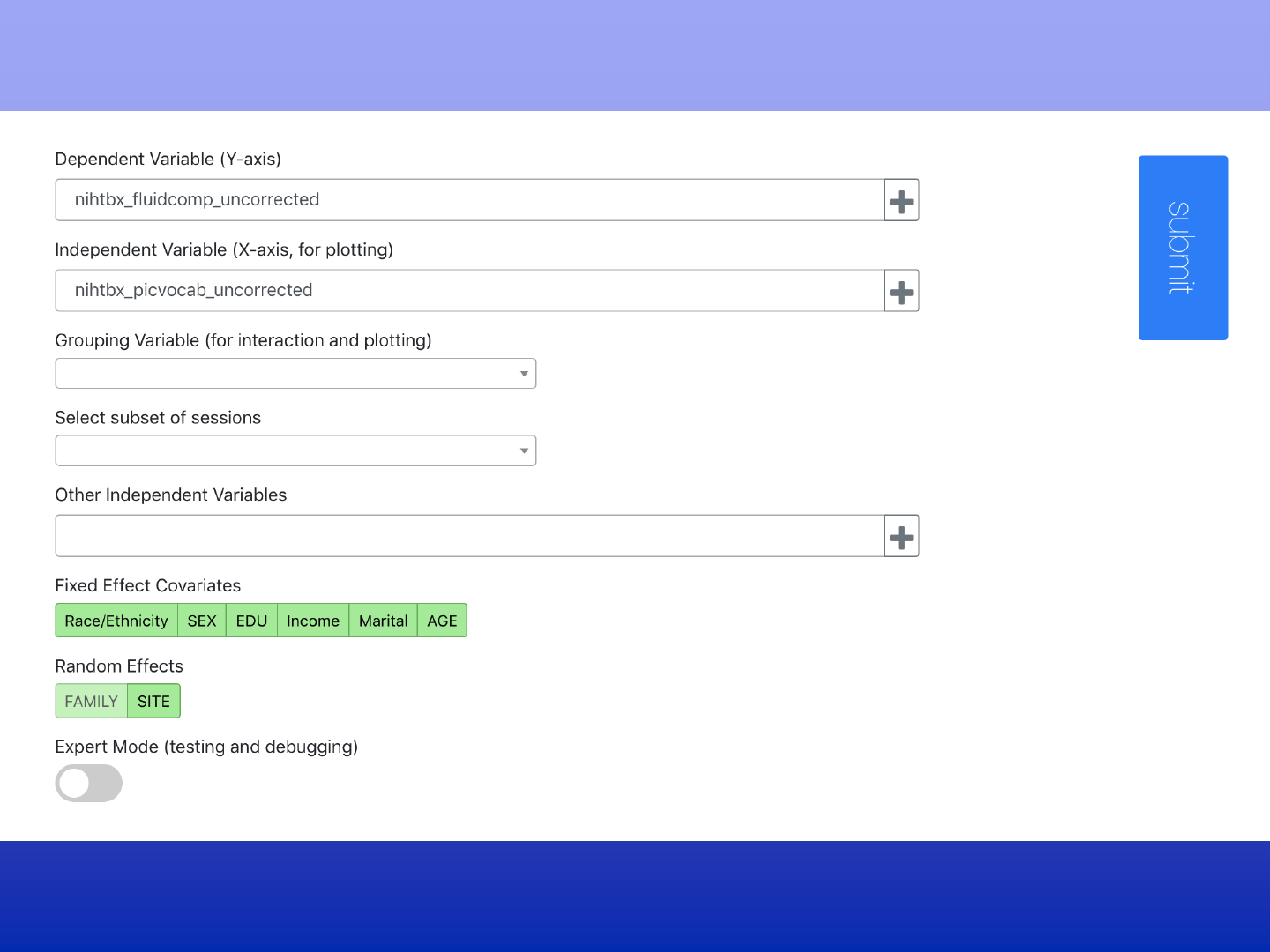
Analyze
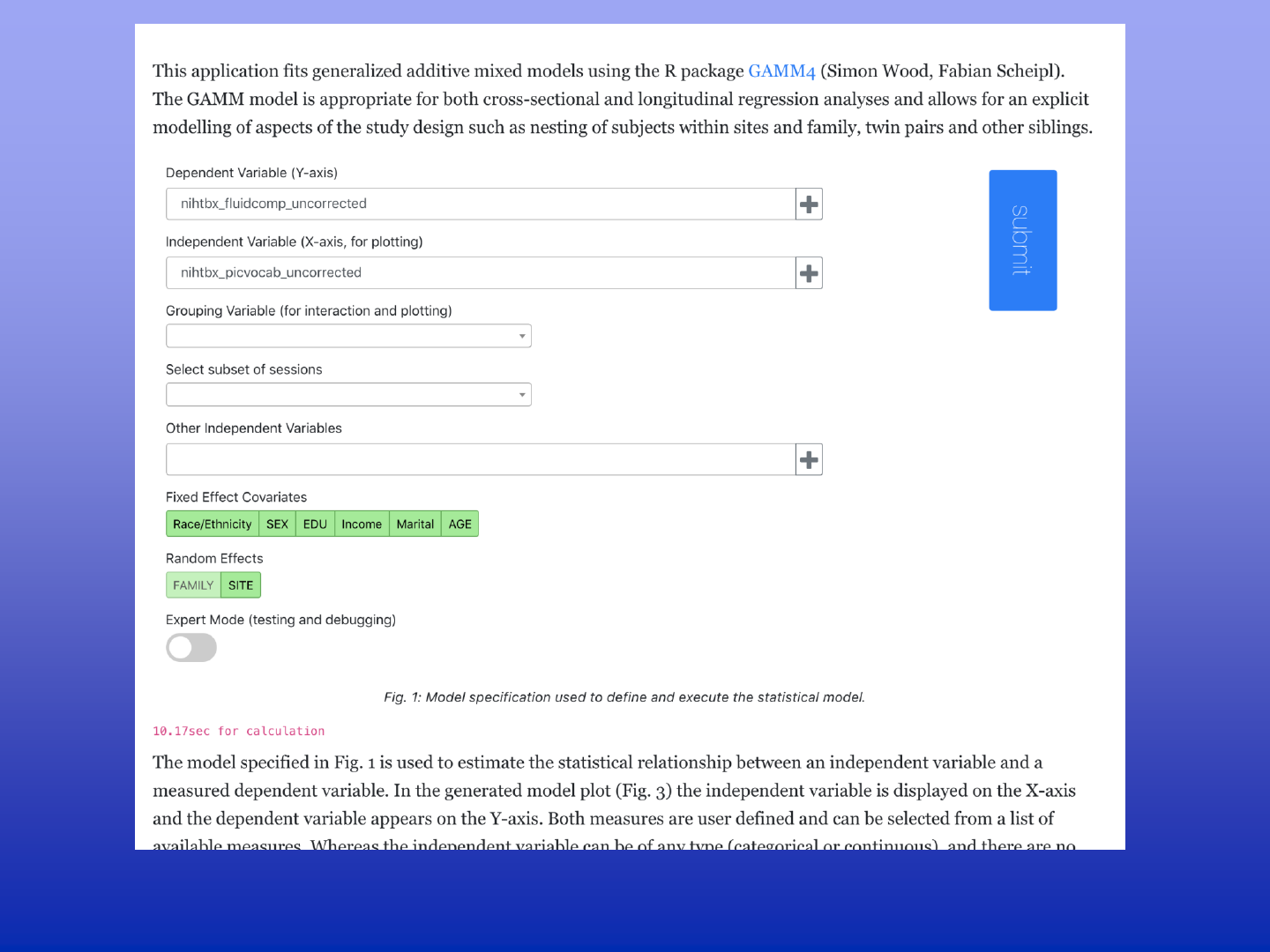
Analysis tutorial mode – expert commentary
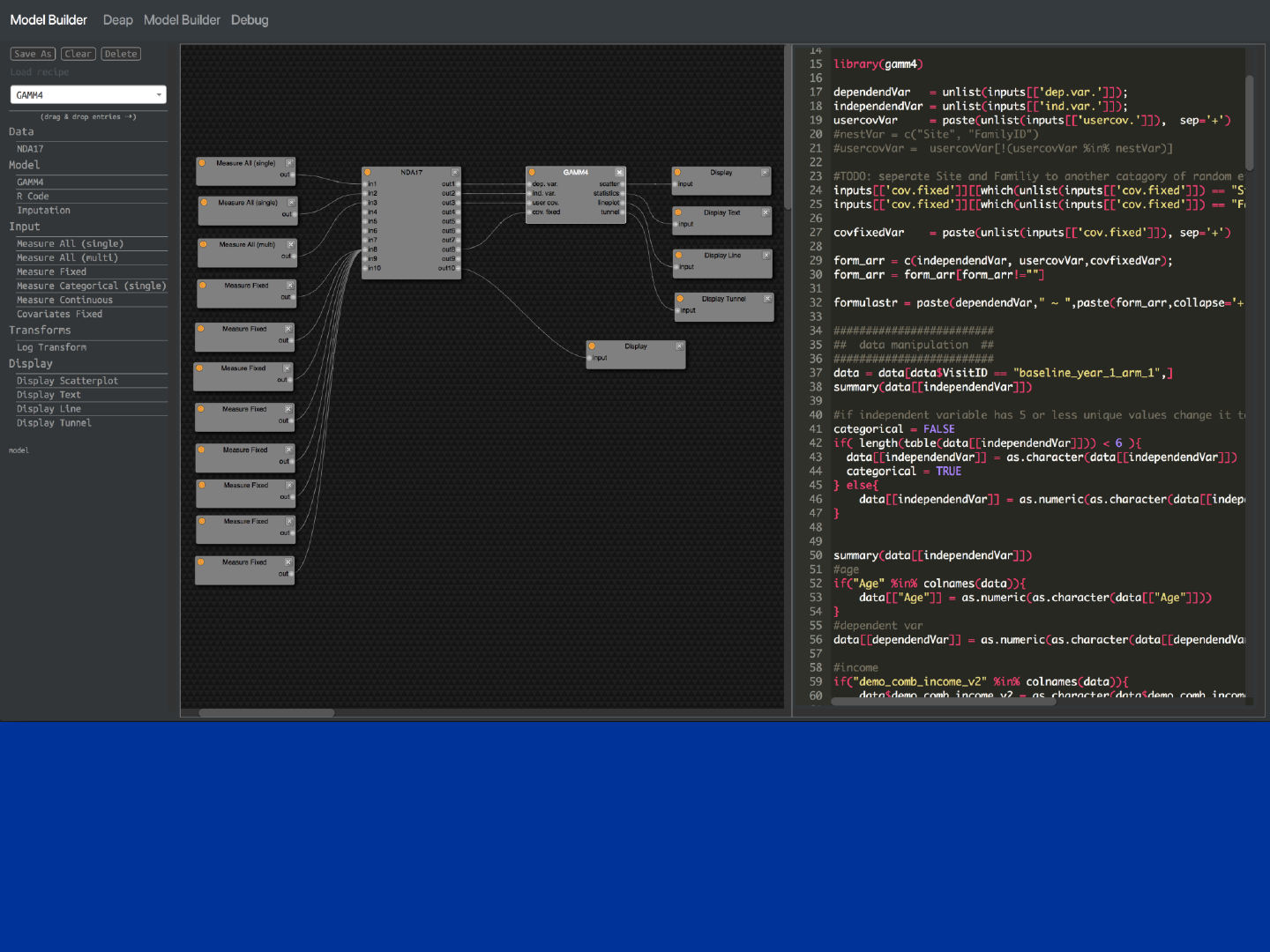
Advanced Usage (Model Builder)
A collaborative environment to integrate advanced statistical analysis features into ABCD. The model builder is
software agnostic. R modules coexist next to python/pandas, Matlab. Data frames are used for inter-nodal
communication. System provides computational cloud resources and each block can be extracted from the system
(data and source-code) for documentation and offline analysis.
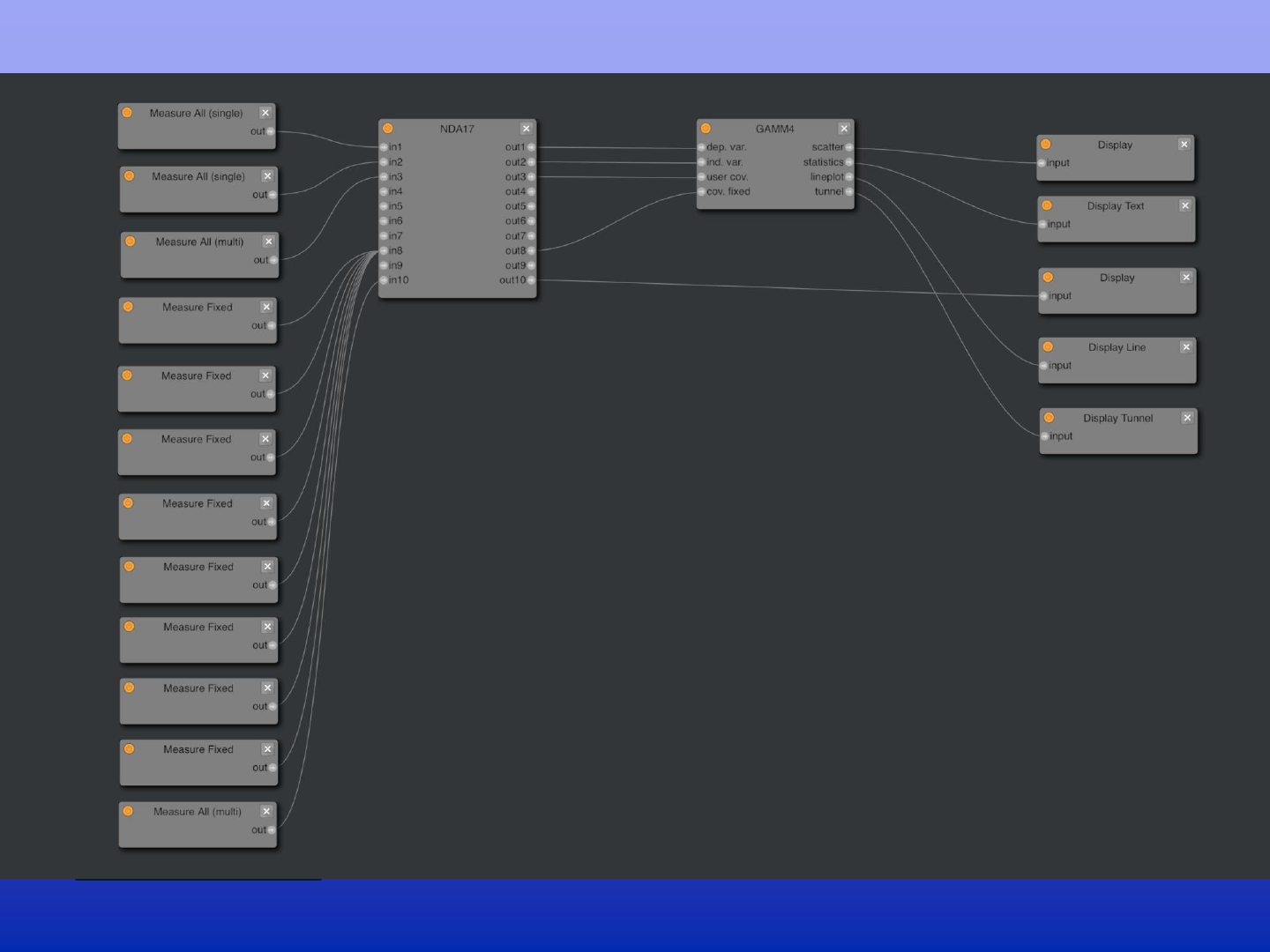
The building blocks for hypothesis testing
Data flow graph (graphical programming) of the Model Builder on DEAP

34
ACKNOWLEDGEMENTS
• The NIMH Data Archive (NDA)
• Greg Farber
• Rebecca Rosen
• Brian Koser
• Trevor Griffiths
• NIH/NIDA
• Gaya Dowling
• Steve Grant
• Elizabeth Hoffman
• Vani Pariyadath
• Anders Dale (PI of the ABCD DAIC: U24 DA041123)
• The DAIC-DEAP Team:
• Hauke Bartsch
• Fangzhou Hu
• Chase Reuter
• ABCD Biostatistics Work Group
